Protein-coding gene in the species Homo sapiens
| SUCLG1 |
|---|
 |
| Identifiers |
|---|
| Aliases | SUCLG1, GALPHA, MTDPS9, SUCLA1, succinate-CoA ligase alpha subunit, succinate-CoA ligase GDP/ADP-forming subunit alpha |
|---|
| External IDs | OMIM: 611224; MGI: 1927234; HomoloGene: 55785; GeneCards: SUCLG1; OMA:SUCLG1 - orthologs |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 2 (human)[1] |
|---|
| | Band | 2p11.2 | Start | 84,423,528 bp[1] |
|---|
| End | 84,460,045 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 6 (mouse)[2] |
|---|
| | Band | 6|6 C1 | Start | 73,225,365 bp[2] |
|---|
| End | 73,253,894 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - kidney tubule
- glomerulus
- metanephric glomerulus
- renal medulla
- jejunal mucosa
- duodenum
- mucosa of transverse colon
- right ventricle
- mucosa of sigmoid colon
- apex of heart
|
| | Top expressed in | - interventricular septum
- digastric muscle
- myocardium of ventricle
- brown adipose tissue
- plantaris muscle
- right kidney
- extensor digitorum longus muscle
- sternocleidomastoid muscle
- extraocular muscle
- vastus lateralis muscle
|
| | More reference expression data |
|
|---|
| BioGPS | 
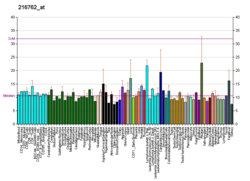 | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - nucleotide binding
- ligase activity
- catalytic activity
- RNA binding
- succinate-CoA ligase (ADP-forming) activity
- succinate-CoA ligase (GDP-forming) activity
- nucleoside diphosphate kinase activity
- GDP binding
- succinate-CoA ligase activity
| | Cellular component | - plasma membrane
- mitochondrion
- mitochondrial inner membrane
- extracellular exosome
- cytosol
- mitochondrial matrix
- succinate-CoA ligase complex (GDP-forming)
| | Biological process | - metabolism
- tricarboxylic acid cycle
- succinyl-CoA metabolic process
- succinate metabolic process
- nucleoside diphosphate phosphorylation
- nucleoside triphosphate biosynthetic process
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | | |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | | |
|---|
| RefSeq (protein) | | |
|---|
| Location (UCSC) | Chr 2: 84.42 – 84.46 Mb | Chr 6: 73.23 – 73.25 Mb |
|---|
| PubMed search | [3] | [4] |
|---|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial is an enzyme that in humans is encoded by the SUCLG1 gene.[5][6]
Structure
The enzyme encoded by SUCLG1 can exist in either a phosphorylated form or a dephosphorylated form. In the dephosphorylated structure, a phosphate ion works in coordination with a histidine residue in the active site and the two alpha helices, one contributed by each subunit of the alphabeta-dimer to stabilize the structure. One of the alpha helices contains amino acids, the modification of which result in conformational changes that accommodate either the bound phosphoryl group or the free phosphate ion.[7]
Function
This gene encodes the alpha subunit of the heterodimeric enzyme succinate coenzyme A ligase. This enzyme is targeted to the mitochondria and catalyzes the conversion of succinyl CoA and ADP or GDP to succinate and ATP or GTP. Mutations in this gene are the cause of the metabolic disorder fatal infantile lactic acidosis and mitochondrial DNA depletion.[8][9]
Clinical significance
Succinate-CoA ligase deficiency is responsible for encephalomyopathy with mitochondrial DNA depletion and mild methylmalonic aciduria. Mutations in SUCLG1 lead to complete absence of SUCLG1 protein and are responsible for a very severe disorder with antenatal manifestations. Furthermore, it is shown that in the absence of SUCLG1 protein, no SUCLA2 protein is found in fibroblasts by western blot analysis. This result is consistent with a degradation of SUCLA2 when its heterodimer partner, SUCLG1, is absent.[10] As mitochondrial DNA depletion in muscle is not a constant finding in SUCLG1 patients, diagnosis should not be based on it; additionally, it may be that alternative physiopathological mechanisms may be considered to explain the combined respiratory chain deficiency observed in these patients.[9]
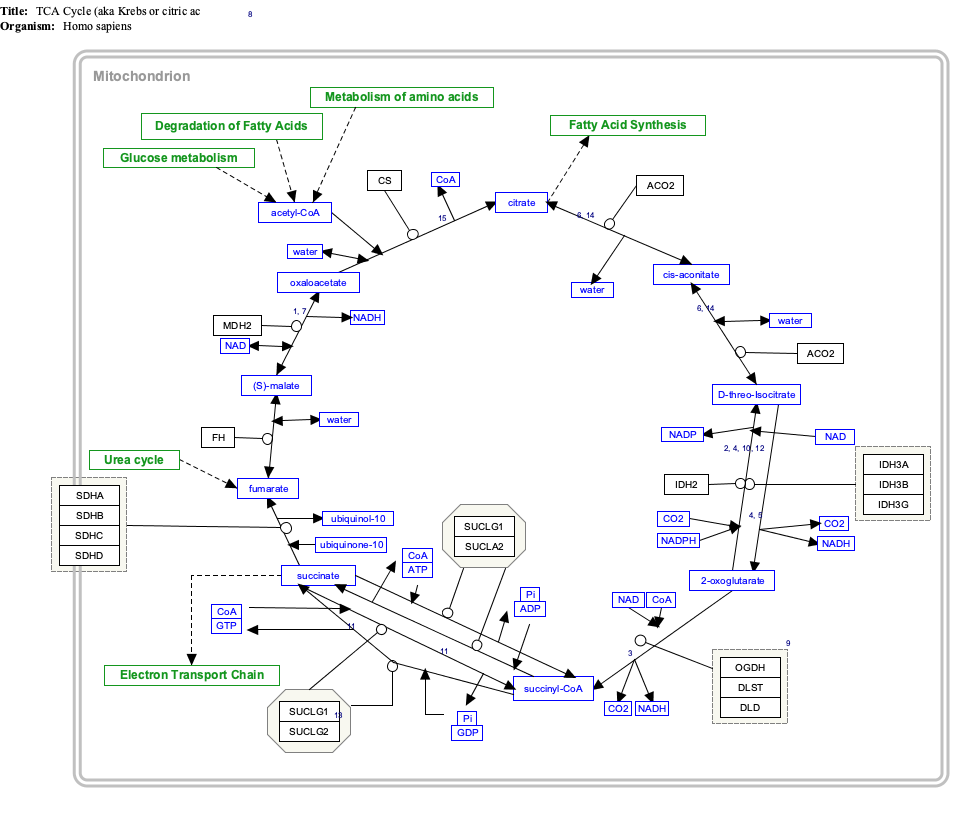
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
[[File:
|alt=TCACycle_WP78 edit]]
- ^ The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78".
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000163541 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000052738 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ James M, Man NT, Edwards YH, Morris GE (Apr 1997). "The molecular basis for cross-reaction of an anti-dystrophin antibody with alpha-actinin". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1360 (2): 169–76. doi:10.1016/s0925-4439(96)00076-2. PMID 9128182.
- ^ "Entrez Gene: SUCLG1 succinate-CoA ligase, GDP-forming, alpha subunit".
- ^ Fraser ME, James MN, Bridger WA, Wolodko WT (Jun 2000). "Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase". Journal of Molecular Biology. 299 (5): 1325–39. doi:10.1006/jmbi.2000.3807. PMID 10873456.
- ^ Ostergaard E (Apr 2008). "Disorders caused by deficiency of succinate-CoA ligase". Journal of Inherited Metabolic Disease. 31 (2): 226–9. doi:10.1007/s10545-008-0828-7. PMID 18392745. S2CID 12722653.
- ^ a b Valayannopoulos V, Haudry C, Serre V, Barth M, Boddaert N, Arnoux JB, Cormier-Daire V, Rio M, Rabier D, Vassault A, Munnich A, Bonnefont JP, de Lonlay P, Rötig A, Lebre AS (Jun 2010). "New SUCLG1 patients expanding the phenotypic spectrum of this rare cause of mild methylmalonic aciduria". Mitochondrion. 10 (4): 335–41. doi:10.1016/j.mito.2010.02.006. PMID 20197121.
- ^ Rouzier C, Le Guédard-Méreuze S, Fragaki K, Serre V, Miro J, Tuffery-Giraud S, Chaussenot A, Bannwarth S, Caruba C, Ostergaard E, Pellissier JF, Richelme C, Espil C, Chabrol B, Paquis-Flucklinger V (Oct 2010). "The severity of phenotype linked to SUCLG1 mutations could be correlated with residual amount of SUCLG1 protein" (PDF). Journal of Medical Genetics. 47 (10): 670–6. doi:10.1136/jmg.2009.073445. PMID 20693550. S2CID 35860287.
Further reading
- Maruyama K, Sugano S (Jan 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (Oct 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Fraser ME, James MN, Bridger WA, Wolodko WT (Jun 2000). "Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA synthetase". Journal of Molecular Biology. 299 (5): 1325–39. doi:10.1006/jmbi.2000.3807. PMID 10873456.
- Suzuki Y, Yamashita R, Shirota M, Sakakibara Y, Chiba J, Mizushima-Sugano J, Nakai K, Sugano S (Sep 2004). "Sequence comparison of human and mouse genes reveals a homologous block structure in the promoter regions". Genome Research. 14 (9): 1711–8. doi:10.1101/gr.2435604. PMC 515316. PMID 15342556.
- Tsang HT, Connell JW, Brown SE, Thompson A, Reid E, Sanderson CM (Sep 2006). "A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex". Genomics. 88 (3): 333–46. doi:10.1016/j.ygeno.2006.04.003. PMID 16730941.
- Ostergaard E, Christensen E, Kristensen E, Mogensen B, Duno M, Shoubridge EA, Wibrand F (Aug 2007). "Deficiency of the alpha subunit of succinate-coenzyme A ligase causes fatal infantile lactic acidosis with mitochondrial DNA depletion". American Journal of Human Genetics. 81 (2): 383–7. doi:10.1086/519222. PMC 1950792. PMID 17668387.
External links
- Overview of all the structural information available in the PDB for UniProt: P53597 (Human Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: O19069 (Pig Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial) at the PDBe-KB.
PDB gallery
-
1euc: CRYSTAL STRUCTURE OF DEPHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE -
1eud: CRYSTAL STRUCTURE OF PHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE -
2fp4: Crystal structure of pig GTP-specific succinyl-CoA synthetase in complex with GTP -
2fpg: Crystal structure of pig GTP-specific succinyl-CoA synthetase in complex with GDP -
2fpi: Crystal structure of pig GTP-specific succinyl-CoA synthetase from polyethylene glycol -
2fpp: Crystal structure of pig GTP-specific succinyl-CoA synthetase from polyethylene glycol with chloride ions |
 | This article on a gene on human chromosome 2 is a stub. You can help Wikipedia by expanding it. |


 1euc: CRYSTAL STRUCTURE OF DEPHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE
1euc: CRYSTAL STRUCTURE OF DEPHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE 1eud: CRYSTAL STRUCTURE OF PHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE
1eud: CRYSTAL STRUCTURE OF PHOSPHORYLATED PIG HEART, GTP-SPECIFIC SUCCINYL-COA SYNTHETASE 2fp4: Crystal structure of pig GTP-specific succinyl-CoA synthetase in complex with GTP
2fp4: Crystal structure of pig GTP-specific succinyl-CoA synthetase in complex with GTP 2fpg: Crystal structure of pig GTP-specific succinyl-CoA synthetase in complex with GDP
2fpg: Crystal structure of pig GTP-specific succinyl-CoA synthetase in complex with GDP 2fpi: Crystal structure of pig GTP-specific succinyl-CoA synthetase from polyethylene glycol
2fpi: Crystal structure of pig GTP-specific succinyl-CoA synthetase from polyethylene glycol 2fpp: Crystal structure of pig GTP-specific succinyl-CoA synthetase from polyethylene glycol with chloride ions
2fpp: Crystal structure of pig GTP-specific succinyl-CoA synthetase from polyethylene glycol with chloride ions


























